DNA: STRUCTURE
DNA, or deoxyribonucleic acid, is the hereditary material in humans and almost all other organisms. Nearly every cell in a person’s body has the same DNA. Most DNA is located in the cell nucleus (where it is called nuclear DNA), but a small amount of DNA can also be found in the mitochondria (where it is called mitochondrial DNA or mtDNA).
DNA is made up of molecules called nucleotides. Each nucleotide contains a phosphate group, a sugar group and a nitrogen base. The four types of nitrogen bases are adenine (A), thymine (T), guanine (G) and cytosine (C). The order of these bases is what determines DNA’s instructions, or genetic code. Similar to the way the order of letters in the alphabet can be used to form a word, the order of nitrogen bases in a DNA sequence forms genes, which in the language of the cell, tells cells how to make proteins. Another type of nucleic acid, ribonucleic acid, or RNA, translates genetic information from DNA into proteins. The entire human genome contains about3 billion bases and about 20,000 genes.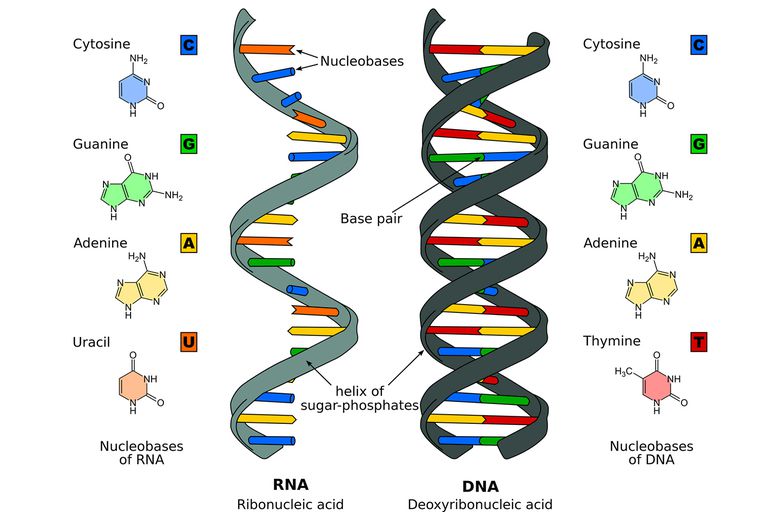
Nucleotides are attached together to form two long strands that spiral to create a structure called a double helix. If you think of the double helix structure as a ladder, the phosphate and sugar molecules would be the sides, while the bases would be the rungs. The bases on one strand pair with the bases on another strand: adenine pairs with thymine, and guanine pairs with cytosine. DNA molecules are long — so long, in fact, that they can’t fit into cells without the right packaging. To fit inside cells, DNA is coiled tightly to form structures we call chromosomes. Each chromosome contains a single DNA molecule. Humans have 23 pairs of chromosomes, which are found inside the cell’s nucleus.
Functions of DNA
DNA usually occurs as linear chromosomes in eukaryotes, and circular chromosomes in prokaryotes. The set of chromosomes in a cell makes up its genome; the human genome has approximately 3 billion base pairs of DNA arranged into 46 chromosomes. The information carried by DNA is held in the sequence of pieces of DNA called genes. Transmission of genetic information in genes is achieved via complementary base pairing. For example, in transcription, when a cell uses the information in a gene, the DNA sequence is copied into a complementary RNA sequence through the attraction between the DNA and the correct RNA nucleotides. Usually, this RNA copy is then used to make a matching protein sequence in a process called translation, which depends on the same interaction between RNA nucleotides. In alternative fashion, a cell may simply copy its genetic information in a process called DNA replication. The details of these functions are covered in other articles; here the focus is on the interactions between DNA and other molecules that mediate the function of the genome.
DNA REPLICATION
Process of DNA replication takes place in following steps:
Fork Formation
Before DNA can be replicated, the double stranded molecule must be “unzipped” into two single strands. DNA has four bases called adenine (A), thymine (T), cytosine (C) and guanine (G) that form pairs between the two strands. Adenine only pairs with thymine and cytosine only binds with guanine. In order to unwind DNA, these interactions between base pairs must be broken. This is performed by an enzyme known as DNA helicase. DNA helicase disrupts the hydrogen bonding between base pairs to separate the strands into a Y shape known as the replication fork. This area will be the template for replication to begin. DNA is directional in both strands, signified by a 5′ and 3′ end. This notation signifies which side group is attached the DNA backbone. The 5′ end has a phosphate (P) group attached, while the 3′ end has a hydroxyl (OH) group attached. This directionality is important for replication as it only progresses in the 5′ to 3′ direction. However, the replication fork is bi-directional; one strand is oriented in the 3′ to 5′ direction (leading strand) while the other is oriented 5′ to 3′ (lagging strand). The two sides are therefore replicated with two different processes to accommodate the directional difference.
Primer Binding
The leading strand is the simplest to replicate. Once the DNA strands have been separated, a short piece of RNA called a primer binds to the 3′ end of the strand. The primer always binds as the starting point for replication. Primers are generated by the enzyme DNA primase.
Elongation
Enzymes known as DNA polymerases are responsible creating the new strand by a process called elongation. There are five different known types of DNA polymerases in bacteria and human cells. In bacteria such as E. coli, polymerase III is the main replication enzyme, while polymerase I, II, IV and V are responsible for error checking and repair. DNA polymerase III binds to the strand at the site of the primer and begins adding new base pairs complementary to the strand during replication. In eukaryotic cells, polymerases alpha, delta, and epsilon are the primary polymerases involved in DNA replication. Because replication proceeds in the 5′ to 3′ direction on the leading strand, the newly formed strand is continuous. The lagging strand begins replication by binding with multiple primers. Each primer is only several bases apart. DNA polymerase then adds pieces of DNA, called Okazaki fragments, to the strand between primers. This process of replication is discontinuous as the newly created fragments are disjointed.
Termination
Once both the continuous and discontinuous strands are formed, an enzyme called exonuclease removes all RNA primers from the original strands. These primers are then replaced with appropriate bases. Another exonuclease “proofreads” the newly formed DNA to check, remove and replace any errors. Another enzyme called DNA ligase joins Okazaki fragments together forming a single unified strand. The ends of the linear DNA present a problem as DNA polymerase can only add nucleotides in the 5? to 3? direction. The ends of the parent strands consist of repeated DNA sequences called telomeres. Telomeres act as protective caps at the end of chromosomes to prevent nearby chromosomes from fusing. A special type of DNA polymerase enzyme called telomerase catalyzes the synthesis of telomere sequences at the ends of the DNA. Once completed, the parent strand and its complementary DNA strand coils into the familiar double helix shape. In the end, replication produces two DNA molecules, each with one strand from the parent molecule and one new strand.
Enzymes involved in the process of DNA replication
DNA replication would not occur without enzymes that catalyze various steps in the process. Enzymes that participate in the eukaryotic DNA replication process include:
DNA helicase: unwinds and separates double stranded DNA as it moves along the DNA. It forms the replication fork by breaking hydrogen bonds between nucleotide pairs in DNA.
DNA primase: a type of RNA polymerase that generates RNA primers. Primers are short RNA molecules that act as templates for the starting point of DNA replication.
DNA polymerases: synthesize new DNA molecules by adding nucleotides to leading and lagging DNA strands.
Topoisomerase or DNA Gyrase: unwinds and rewinds DNA strands to prevent the DNA from becoming tangled or supercoiled.
Exonucleases: group of enzymes that remove nucleotide bases from the end of a DNA chain.
DNA ligase: joins DNA fragments together by forming phosphodiester bonds between nucleotides.
RNA stands for ribonucleic acid. It is an important molecule with long chains of nucleotides. A nucleotide contains a nitrogenous base, a ribose sugar, and a phosphate. Just like DNA, RNA is vital for living beings.RNA (ribonucleic acid) is a nucleic acid polymer where the carbohydrate is ribose. RNA is generally single-stranded, as DNA is transcribed by RNA polymerases into mRNA (messenger RNA), which is read by ribosomes to generate protein (translation). Biologically active RNAs, including transport, ribosomal and small nuclear RNA (tRNA, rRNA, snRNAs) fold into unique structures guided by complementary pairing between nucleotide bases.